Before running your own analysis using the ADMIRE web service, you should have the following files on your local desktop:
- a SampleSheet.csv file and
- file directories named after the assays Sentrix-ID containing two *.idat files per sample.
To reduce the amount of data to send to the web server, compress the folders containing the *.idat files. To do this, select all folders and use built-in solutions from Windows or Mac to create a file that ends with
- .tar.gz, .tgz - uses the tar compression together with gzip or
- .tar.bz2, .tbz2, .tar.bzip2
Now register a user by selecting the User tab in the top panel of the ADMIRE web page, followed by a click on Register. Fill in your email address and choose a password and a public name. You don't have to wait for a confirmation email and can work right out of the box.
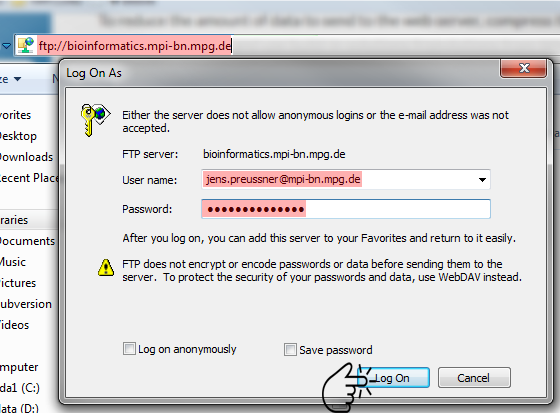
By creating a user account, you'll also be granted access to your personal FTP space on the server. As a next step, upload the compressed idat files into your remote FTP space. To do so, use any FTP client or Windows Explorers built-in capabilities to connect to the FTP server at ftp://bioinformatics.mpi-bn.mpg.de:
Now drag and drop the SampleSheet.csv and the compressed file you created earlier into the FTP space:
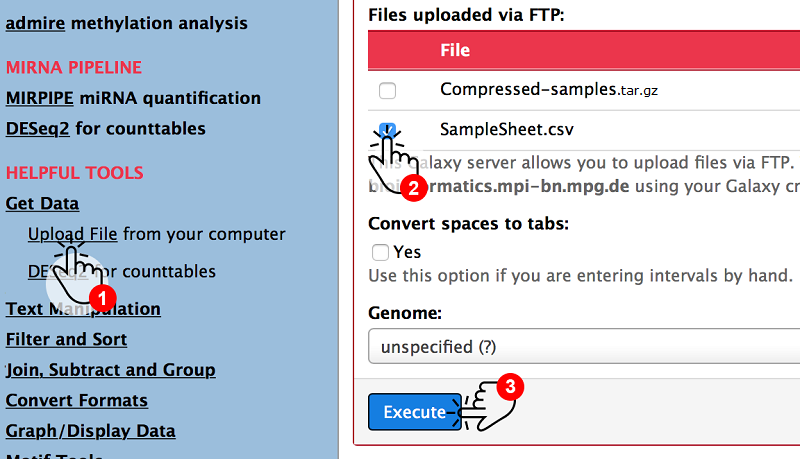
Change back to the web server and use the Upload Tool (1) to select the SampleSheet.csv file (2) only and upload it by clicking on Execute (3).
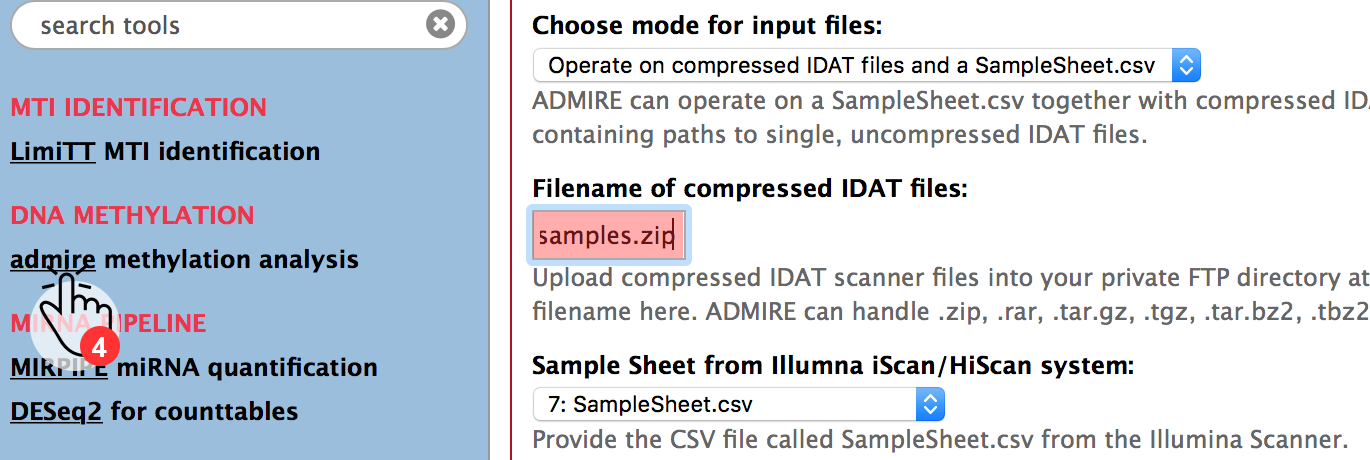
Now choose the ADMIRE tool from the toolbar (4) and fill in the file name of the compressed idat files.
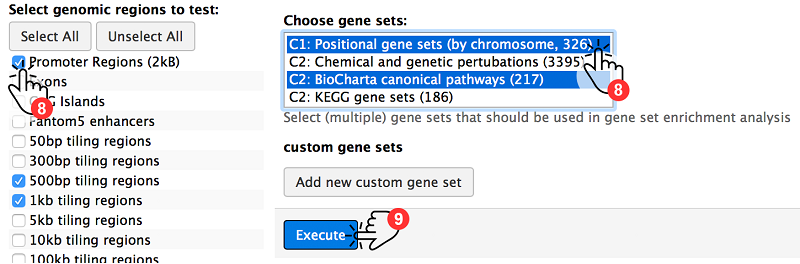
As a last step, choose genomic regions of interest and (optional) gene sets (8). Hit the Execute button to see ADMIRE in action (9).